 Graphical Abstract: RSC
Graphical Abstract: RSCAbstract
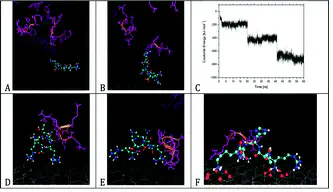
Investigating the adsorption of peptides on inorganic surfaces, on the molecular level, is fundamental for medicinal and analytical applications. Peptides can be potent as linkers between surfaces and living cells in biochips or in implantation medicine. Here, we studied the adsorption process of the positively charged pentapeptide RTHRK, a recently identified binding sequence for surface oxidized silicon, and novel analogues thereof to negatively charged mica surfaces. Homogeneous formation of monolayers in the nano- and low micromolar peptide concentration range was observed. We propose an alternative and efficient method to both quantify binding affinity and follow adhesion behavior. This method makes use of the thermodynamic relationship between surface coverage, measured by atomic force microscopy (AFM), and the concomitant free energy of adhesion. A knowledge-based fit to the autocorrelation of the AFM images was used to correct for a biased surface coverage introduced by the finite lateral resolution of the AFM. Binding affinities and mechanisms were further explored by large scale molecular dynamics (MD) simulations. The combination of well validated MD simulations with topological data from AFM revealed a better understanding of peptide adsorption processes on the atomistic scale. We demonstrate that binding affinity is strongly determined by a peptide’s ability to form salt bridges and hydrogen bonds with the surface lattice. Consequently, differences in hydrogen bond formation lead to substantial differences in binding affinity despite conservation of the peptide’s overall charge. Further, MD simulations give access to relative changes in binding energy of peptide variations in comparison to a lead compound.